
Key Points:
•Macroevolution is term for evolutionary changes recognized above the species level.
•One major macroevolutionary pattern testable in the fossil record is whether species-level change is mostly gradual (phyletic gradualism) or in short bursts separated by long period of stasis (punctuated equilibrium).
•Under phyletic gradualism, then species don't have real beginnings or ends, and are just "chronospecies" with anagenetic origins; under the latter, the case can be made that species are "real" rather than purely arbitrary.
•In recent decade it has been recognized that evolution is highly controlled by development: this is represented by the new study of "evo-devo".
"Naturalists try to arrange the species, genera, and families in each class, on what is called the Natural System. But what is meant by this system? Some authors look at it merely as a scheme for arranging together those living objects which are most alike, and for separating those which are most unlike; or as an artificial means for enunciating, as briefly as possible, general propositions, that is, by one sentence to give the characters common, for instance, to all mammals, by another those common to all carnivora, by another those common to the dog-genus, and then by adding a single sentence, a full description is given of each kind of dog. The ingenuity and utility of this system are indisputable. But many naturalists think that something more is meant by the Natural System; they believe that it reveals the plan of the Creator; but unless it be specified whether order in time or space, or what else is meant by the plan of the Creator, it seems to me that nothing is thus added to our knowledge. Such expressions as that famous one of Linnaeus, and which we often meet with in a more or less concealed form, that the characters do not make the genus, but that the genus gives the characters, seem to imply that something more is included in our classification, than mere resemblance. I believe that something more is included; and that propinquity of descent, the only known cause of the similarity of organic beings, is the bond, hidden as it is by various degrees of modification, which is partially revealed to us by our classifications." -- Chapter 13, On the Origin of Species by Means of Natural Selection (1859), Charles Darwin
"The affinities of all the beings of the same class have sometimes been represented by a great tree. I believe this simile largely speaks the truth. The green and budding twigs may represent existing species; and those produced during each former year may represent the long succession of extinct species. At each period of growth all the growing twigs have tried to branch out on all sides, and to overtop and kill the surrounding twigs and branches, in the same manner as species and groups of species have tried to overmaster other species in the great battle for life. The limbs divided into great branches, and these into lesser and lesser branches, were themselves once, when the tree was small, budding twigs; and this connexion of the former and present buds by ramifying branches may well represent the classification of all extinct and living species in groups subordinate to groups. Of the many twigs which flourished when the tree was a mere bush, only two or three, now grown into great branches, yet survive and bear all the other branches; so with the species which lived during long-past geological periods, very few now have living and modified descendants. From the first growth of the tree, many a limb and branch has decayed and dropped off; and these lost branches of various sizes may represent those whole orders, families, and genera which have now no living representatives, and which are known to us only from having been found in a fossil state. As we here and there see a thin straggling branch springing from a fork low down in a tree, and which by some chance has been favoured and is still alive on its summit, so we occasionally see an animal like the Ornithorhynchus or Lepidosiren, which in some small degree connects by its affinities two large branches of life, and which has apparently been saved from fatal competition by having inhabited a protected station. As buds give rise by growth to fresh buds, and these, if vigorous, branch out and overtop on all sides many a feebler branch, so by generation I believe it has been with the great Tree of Life, which fills with its dead and broken branches the crust of the earth, and covers the surface with its ever branching and beautiful ramifications." -- Chapter 4, On the Origin of Species by Means of Natural Selection (1859), Charles Darwin
SYSTEMATICS
The diversity of living things presents us with a seemingly infinite variety. The science of systematics is dedicated to identifying and ordering the diversity of living things. Since prehistory, systematists have employed taxonomic systems in which organisms are classified into groups or taxa (singular: taxon). Many different taxonomic systems are conceivable, but all have the following features:

For example, in our lives, we have all employed the taxonomic system in which animals are classified according to the organizational principle of their utility to humans. Generally, there is little ambiguity.
Livestock |
Pet
|
Vermin |

We have already looked at the grammar of higher taxa in the Linnaean system. But what do these higher taxa mean, and especially what was the significance (if any) of the traditional ranks (Family, Order, etc.)?
For a long time people treated taxa given equivalent ranks AS equivalent in some meaningful way. For instance, Van Valen's Red Queen hypothesis was originally conceived by testing to see if families had their chances of survival increase with longer survivorship, as if a family of mollusks were equivalent to a family of brachiopod or vertebrate.
But consider that the ants are traditionally considered simply as one family (Formicidae) with more than 12,000 species. In contrast the Class (a rank two levels higher) Mammalia has a mere 5450 species, and within it the Order (still a rank higher than family) Tubulidentata has only one living species! So clearly taxa of the same rank do not have equivalent number of species, or of morphological disparity, nor age of origin. In fact, they are merely book-keeping structures.
For that purposes that is fine. But if your goal is to discover evolutionarily significant groups, ranks do us no good. (And in point of fact taxonomists long ago found the need to interject super-, sub-, infra-, and other prefix-versions of ranks (superclass, infraorder, subfamily, etc.) in order to chart out recognizable divisions of the diversity of life.
Although the ranks might not be real, the actual named taxa were useful.
Linnaeus's system gave an important aspect to compare groups: the taxonomic level at which any two species were united. So lions (Panthera leo) and jaguars (Panthera once) were united at the low level of the genus Panthera; lions and cheetahs (Acinonyx jubatus) were united at the higher level of the family Felidae; lions and bears (Ursidae) at the level of the order Carnivora; and so forth.
But what did it really mean by "more closely related"? Darwin (see first quote at top) identified an answer: our classifications were pointing at the degree of recency of common ancestors ("propinquity of descent", in Victorian English). So species within a genus had only recently diverged from each other; species united at the level of a family had more distant ancestors; and so on.
Note: something that Darwin also noted, even before he wrote The Origin, is that the idea of "higher" and "lower" was an outdated concept. As all living creatures have equally ancient ancestry, nothing alive is really any more or less "high".
Consequently, THERE IS NO EVOLUTIONARY SCALE! Nothing is "higher" or "lower" on an "evolutionary scale". There IS "primitive" (closer to the ancestral condition) and "derived" (more transformed or specialized from the ancestral condition), but these are relative and specific statements. And they might apply differently to different organ systems in the same pairs of organisms. For instance, humans have vastly more derived (transformed) brains relative to star-nosed moles, but the mole's nose is vasty more advanced compared our primitive one.
Darwin observed (second quote at top) that the primary pattern to related the various species past and present is a Tree of Life:
(Darwin wrote in a notebook that a better metaphor would be the "coral of life", since anything below the tips is dead material made of rock: i.e., fossils.)
The Tree of Life would represent the basis for the higher level classifications: the farther down the tree any two branches joined up, the farther back in time was the divergence between those branches and the more distantly related they were.
Linnaean method continued after Linnaeus' death. Looked at the diversity of life, and grouped organisms together because of shared similar traits
Ernst Haeckel coined the word phylogeny for a "family tree" of Life (or some subset thereof). This is a representation of an hypothesis of the actual historical pattern of ancestors and descendants. Since all living things are related (at least at some level), there should be one true historical branching Tree of Life. The task, then, is to reconstruct it.
Ideally, one could combine information from living creatures with their ancestors to complete this tree. But there are difficulties:
PHYLOGENETIC SYSTEMATICS
But how could we reconstruct the shape of the Tree of Life? Evolutionary biologists from the mid-19th Century until the later 20th Century lacked a good repeatable methodology to establish this. The most common method was simply arranging groups of living and extinct species which seemed to be closely related based on their anatomical features on a stratigraphic scale, and connecting the earlier forms with later ones on the assumption that they were the direct ancestors.
The problem, as we noted last lecture, is that being an ancestor is a very, very specific thing. Being an early relative is not enough: an ancestor is specifically a great great great… grandparent, not a cousin a few dozen times removed.
How to deal with these issues? A solution was found by East German dipterist (fly specialist) Willi Hennig in the 1950s (but not know in English until the 1960s). The technique was called phylogenetic systematics, or more commonly cladistics (from the Greek "klados" ["branch"] for clade, meaning a branch of the Tree of Life). Hennig recognized that finding direct ancestors in the fossil record would be hard, and demonstrating that the fossil was a direct ancestor rather than a close relative of that ancestor would be even harder. So instead, his method focused on estimating the pattern of shared common ancestry. In order to make that estimation, cladistics looks at the distribution of derived characters [evolutionary specializations]. Here is how it works. Below is a grouping of some modern animals:
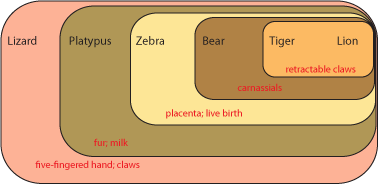
Each set contains multiple taxa. Each set is characterized by derived characters not seen in the taxa outside that set. For example, the lizard shares with all the others the presence of a five-fingered hand and claws. The platypus and all the others except the lizard shares fur and milk. Since lizards (and alligators and frogs and fish and many other animals not shown here) do not have fur and milk, it is most likely explained by having evolved in the common ancestor of platypus, zebra, bear, tiger, and lion AFTER that ancestor had diverged from the common ancestor of lizard, platypus, zebra, bear, tiger, and lion. In other words, platypus, zebra, bear, tiger, and lion most likely had a more recent common ancestor with each other than any did with lizards.
It is generally easier to show this pattern using a simplified stick figure called a cladogram than the set diagram. Below is the cladogram showing the same information as the sets above:
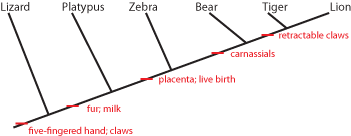
The little red "tick marks" show that the derived character state appears at that point of the cladogram, and is passed on upwards. To show what that means, see the following:

So the common ancestor of zebra, bear, tiger, and lion evolved a placenta and live birth, and passed it on to its descendants. And the common ancestor of bear, tiger, and lion (but NOT zebra) had its molars evolve into carnassials (shearing teeth), and passed that trait on.
Not all character states are useful in figuring out evolutionary relationships. Primitive (or ancestral) states do not help. For example, both lizard and platypus lay shelled eggs rather than have a placenta and live birth. Does that mean that these two animals shared a more recent common ancestor with each other than either did with the other animals here? NO! In fact, having a shelled egg is the ancestral state for a larger (more inclusive) group of animals, including birds, crocodilians, turtles, snakes, echidnas, etc. Having a shelled egg is simply the condition (state) of all advanced terrestrial vertebrates. The technical term for a shared primitive character is a symplesiomorphy.
However, fur and milk are derived states not found in terrestrial vertebrates other than mammals, and indicate a common ancestor to these animals not found in lizards, birds, crocodilians, turtles, etc. The technical term for a shared derived character is a synapomorphy
Similarly, sharing a five-fingered hand doesn't unite the lizard, platypus, and bear (for example) exclusive of the zebra. A five-fingered hand is a symplesiomorphy, and will be passed on that way unless it evolves into something else.
(NOTE: a character can be "primitive" at one level, but "derived" in a different context. In this cladogram, retractable claws is a derived feature that shows us that lions and tigers share a more recent common ancestor with each other than with anything else on this cladogram. However, if we were trying to figure out the relationship between lions, tigers, jaguars, and leopards, than retractable claws would be a primitive charcter shared by all of them, and so useless.)
Zebras have only a single finger on each hand. This is an autapomorphy or unique derived character. Although autapomorphies might be useful in understanding how that animal lived, they don't help us figure out where they fit in the cladogram, since those traits evolved after the ancestor of (in this case) zebras diverged from any shared ancestor of any of the other animals on the list. (However, if we added horses or donkeys to this cladogram, a single finger on each hand would be a good shared DERIVED character, and help us figure out that horses and donkeys share a common ancestor not shared with bears, lions, tigers, etc.).
Convergent characters could actually mislead us. For example, tigers and zebras both have stripes, whereas the other animals on this list don't. Does that mean that tigers and zebras share a common ancestor not shared by the other animals? We might think so at first, but the shared presence of carnassials in bears, lions, and tigers (but not zebras) and of retractable claws in lions and tigers (but not zebras) indicate a different pattern of ancestry. Convergent characters can be very annoying, because they might cause us to make the wrong estimation of the cladogram.
An additional type of character (not seen here) are reversals: when an ancestor had a derived condition, but the trait evolved "back" to the primitive condition. For example, snakes are descendants of typical legged lizards. But snakes have evolved the loss of limbs, so that they resemble the limbless ancestor of vertebrates. For another, while the common ancestor of living birds could fly, but ostriches, rheas, cassowaries, etc. have reverted to the primitive flightless state. Reversals that are unique characters do not help in estimating evolutionary relationships, but ones which are shared might. In general, though, reversals (like convergent characters) are annoying and are a major source of error for our analyses. Collectively, we call convergent characters and reversals homoplasies.
So, in review:
How do we tell which states (conditions) of a character are ancestral, and which are derived? One common method is to look at outgroups: more distant relatives of the taxa in question. Looking at fish, amphibians, turtles, lizards and snakes, crocodilians, and birds show that no placenta is the primitive condition among vertebrates, and that the placenta of some mammals is a derived condition. (It is best to look at multiple outgroups.)
Let's see how this works in a simple cladistic analysis of some imaginary beetles. We assume that they are related somehow, but we don't know if B shares a more recent common ancestor with C or A, or if C and D are more closely related to one another than to B.

The first thing we do is notice how they are different
| Character 1. Large jaws present 2. Small antennae present 3. Spots present 4. Stripes present |
A 0 0 0 0 |
B 1 0 0 0 |
C 1 1 1 0 |
D 1 1 1 1 |
This matrix records whether the observed state for each taxon is ancestral or derived. How do we know? You may have noticed that we we haven't had much to say about A. In this analysis, A is the outgroup taxon. This is a beetle that, on the basis of some prior information, we can assume is more distantly related to beetles A, B, and C than any of them are to one another. Maybe we found it fossilized in amber. The outgroup is our standard for what is derived and what isn't, in that anything we see in it, we assume to be the ancestral state. Incidentally, because it has smaller mandibles than the others, I've included a "large mandible" characteristic in the matrix.
Tree 1:

Tree 2:

Tree 3:

We now compare every possible evolutionary tree arrangement by mapping onto them the simplest possible distribution of each character state change.
The evolutionary novelty of large mandibles, for example, can be explained most simply assuming that it appeared once before the last common ancestor of A, B. and C, who simply inherited it. Short antennae are a different story. The outgroup has long antennae, so we assume this is the ancestral state, but beetle C retains them also. In some tree arrangements, this implies that short antennae they evolved independently in A and B. After mapping the character changes onto the trees, we count them up. Tree 1 implies a minimum of six changes, tree 2 suggests a minimum of eight and tree 3 would require a minimum of seven. Each of these arrangements is a plausible hypothesis of evolutionary history. Now, we must choose the best hypothesis. To do that, we use the principle of parsimony, which holds that the simplest solution is usually the best. For us, the simplest hypothesis is the one that invokes the fewest character state changes. That is tree 1, with only six changes. Is this the true tree? We do not know. the best we can say is that it is our best approximation based on current knowledge.
The above example uses the classic method of phylogeny reconstruction: Maximum Parsimony. MP analyses optimize on finding the tree requiring the smallest number of evolutionary transitions. This was computationally relatively simple to do, and could be easily implemented no matter what kind of data (morphological, genetic, genomic, etc.) data were used. However, MP analyses have some difficulties. In cases of large database size, there is a tendency to recover false positive associations due to what is called long-branch attraction (basically, homoplastic signals overriding the synapomorphic ones.)
For this (and various other operational and mathematical) reason, in the last decade or two there has been a move to use Bayesian methods of phylogeny reconstruction. These algorithms are based on probability and likelihood. For statistical reasons they are less subject to long-branch attraction. And they are often more precise (that is, more likely to result in more resolved trees.)
But are Bayesian analyses more accurate? That is, are they better at recovering the ACTUAL real phylogeny that existed in the History of Life? That remains an unresolved question. Every month or so a paper comes out showing one or the other method is better at resolving trees under some set of circumstances. But at present nothing has come up that definitively and decisively shows that either method is actually superior. Most systemicists today prefer to use both methods and present the results from both. (Among other things, the results shared by both trees are very likely well-supported.) In the end, remember that a cladogram is an hypothesis, subject to future evaluation and refutation.
Of course, an alert observer would raise a serious question: Phylogenetic Systematics absolutely requires us to identify variation in homologous structures, genes, or behaviors. How do we know when character states are homologous? Cladists rely on three tests to falsify hypotheses of homology:
No fossil is complete, so we can't always directly tell what character state was present in a given species. But we
can sometimes us the phylogenetic position of a fossil to help us estimate what that state was. Below is a cladogram
of tyrannosauroids (tyrant dinosaurs), with the number of fingers on the hand listed:
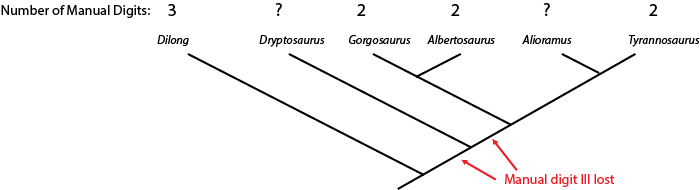
The hand is not yet known for Alioramus or Dryptosaurus. However, we know that Dilong has the
ancestral condition of a three fingered manus (shared with many other carnivorous dinosaur groups), and that Gorgosaurus,
Albertosaurus, and Tyrannosaurus all share the evolutionary specialization "loss of manual digit III" (meaning
that they had a two-fingered hand). The simplest explanation is that the concestor of these three dinosaurs had already lost
manual digit III, and passed on this derived trait to the different descendant lineages.
Because Alioramus (based on other evidence) is ALSO a descendant of that concestor, we predict that it too had a two fingered hand. To assume it had a three fingered hand would mean assuming an evolutionary change (in this case, a reversal) for which we had no direct evidence. The simplest explanation for our current observations is that Alioramus was a two-fingered dinosaur.
But things are unclear for Dryptosaurus. It might be that the loss of manual digit III occured after the ancestor of Dryptosaurus diverged from the concestor of Alberosaurus through Tyrannosaurus: if that were the case, Dryptosaurus would have a three fingered hand. OR it might be that the loss of manual digit III happened before the Dryptosaurus lineage and the Albertosaurus + Tyrannosaurus lineage had diverged: in that case, Dryptosaurus was a two fingered dinosaur. So at present, things remain ambiguous for this taxon.
If we did have to choose, however, there are two different alternative strategies (or optimizations) for making this decision. Under the accelerated transformation (ACCTRANS) optimization, we assume the earliest possible parsimonious transformation (and thus emphasize gains and losses rather than convergence). Under the ACCTRANS model, Dryptosaurus would be optimized as having the two-fingered hand state. Alternatively, there is the delayed transformation (DELTRANS) optimization, in which we assume the latest possible parsimonious transformation (and thus emphasize convergence over gains and losses). Under DELTRANS, Dryptosaurus would be optimized as having a three-fingered hand state.
Sometimes, this can make a significant difference. Consider the example below:
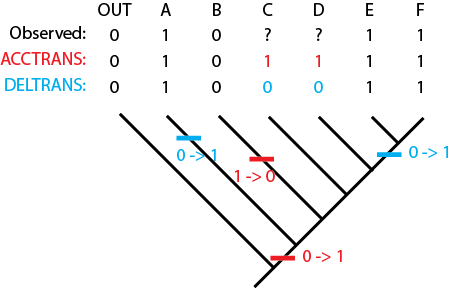
Under ACCTRANS we infer that the concestor of the entire ingroup has the derived condition, and that B is the oddball in reverting to the ancestral condition. But under DELTRANS the clade overall is inferred to have state O, and the divergent members A and E+F independently evolved this trait. So we would have two radically different alternative models of the evolution of this feature and the typical nature of this clade.
The Extant Phylogenetic Bracket
For any fossil form, we can find the two closest outgroups which are still extant. These represent the extant phylogenetic bracket (or EPB)
of the fossil taxon. If we hypothesize that the extinct form has a particular soft tissue structure, behavior, or other character that
cannot be directly observed from the fossil record, this inference falls in three categories of decreasing confidence:
Below are examples of each of these:

In the first case, the basal horse Eohippus
is bracketed by Ceratomorpha (rhinos plus tapirs) and Equus (the living horse genus). Both of the EPB possess fur, so the inference
that Eohippus was furry is a secure Type I inference.
In the second case, the therapsid "protomammal" Thrinaxodon is bracketed by living mammals (which possess fur) and living reptiles (which do not). We might infer that Thrinaxodon was furry, but this is a less-secure Type II inference. Of course, preservation in a Konservat-Lagerstatt might someday show that Thrinaxodon had fur (direct fossil evidence) OR there might be a similar discovery of fur in a taxon more distantly related to mammals than Thrinaxodon (which would make the furry Thrinaxodon hypothesis a Type I inference, with an extinct but well-preseved taxon sitting in as part of the bracket).
In the final case, some paleontologists have hypothesized that the bird-hipped herbivorous dinosaurs in Ornithischia had mammal-like cheeks. While this is argued on circumstantial evidence, it represents the least-secure Type III inference using the EPB method, as neither members of the EPB (crocodylians and birds) have cheeks.
Consensus and Tree Support
Because of missing information, convergence, and reversals, there can be confusion as to the actual pattern of the Tree of
Life. It is often the case that multiple different cladograms are equally well supported ("equally parsimonious"). In
those cases, we can list all of them separately, or we can show a "consensus cladogram" that shows the ambiguity. This is
seen by the presence of a polytomy: a node with more than two branches coming out of it. Below are three different
equally simple cladograms for the ornithischian (bird-hipped) dinosaurs, and the consensus cladogram below:
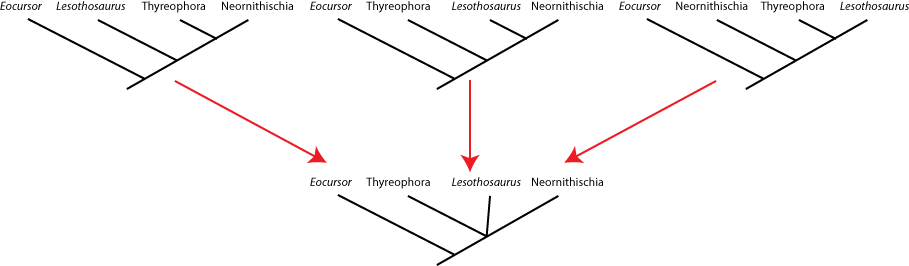
Note the polytomy in the consensus cladogram.
There are several different strategies for generating consensus trees. Each maximizes the information in a different way, but like
map projections of a globe onto two dimensions, they all lose or distort some element
of the data in some fashion. Here are some of the main types of consensus trees:
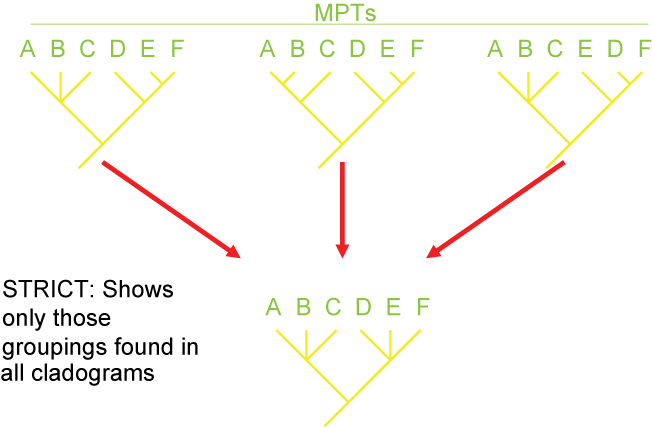
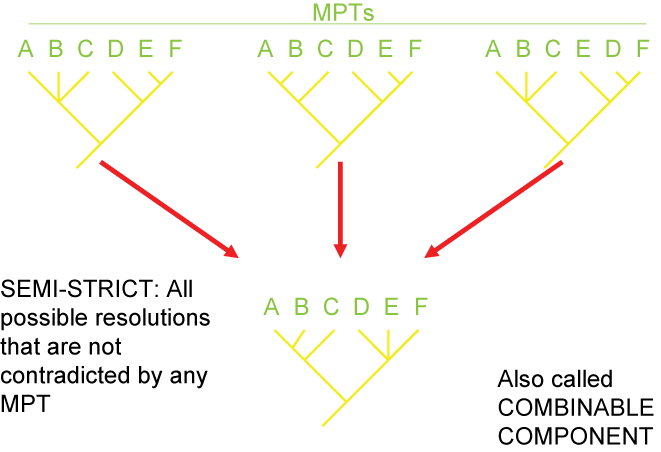
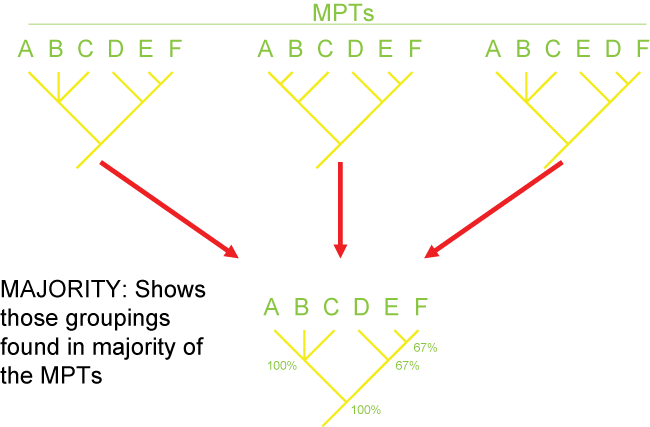
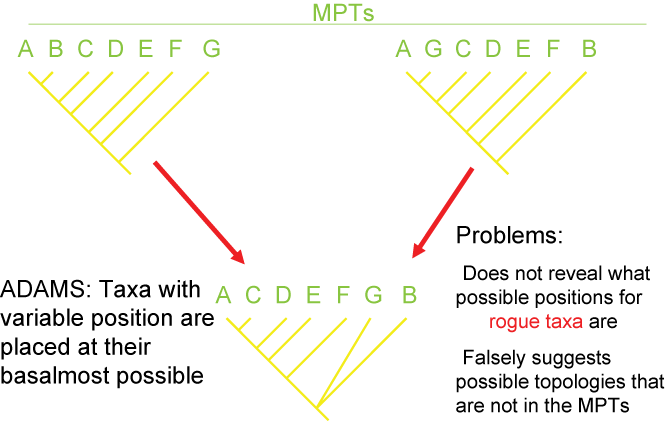
Additionally, there are different methods for evaluating how secure the different results (i.e., the nodes generated) of the analysis are. The
two main methods are Bremer Support (also called decay index):
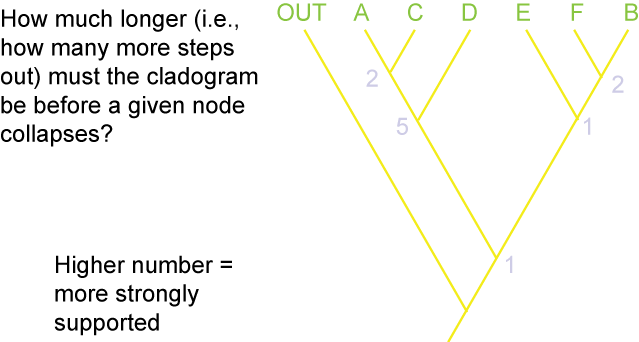
and bootstrap value:
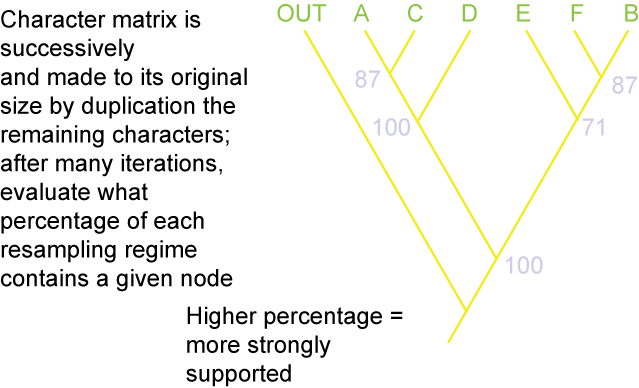
Paleontologists and other biologists have started to use phylogenetic definitions for taxa: that is, they specify a particular relationship and any taxon that qualifies under that relationship is member of that clade. Biologists have coined the word concestor for most recent common ancestor. While the chances of finding the concestor of any two (or more) clades is slim, we can recognize that there did exist such a species somewhere in time. This recognition gives us a way of defining taxa.
Paleontologists sometimes find it useful to refer to the crown-group: that is, the clade formed by all descendants of concestor of the living members of a clade. The total-group is the clade comprised of the crown-group plus all taxa sharing a more recent common ancestor with it than with the next most closely related crown-group. Finally, the stem-group is the paraphyletic part of the total group excluding the crown group. In the example below, the total-group Cephalopoda includes the modern Nautilida and Coleoidea and all taxa more closely related to them than to Scaphopoda. Nautilida and Coleoidea define the crown-group, which also contains the extinct clade Ammonoidea (which although extinct is nevertheless a descendant of the concestor of nautilids and coleoids.) Plectronocerida and Endocerida, however, belong to the stem-group of cephalopods.
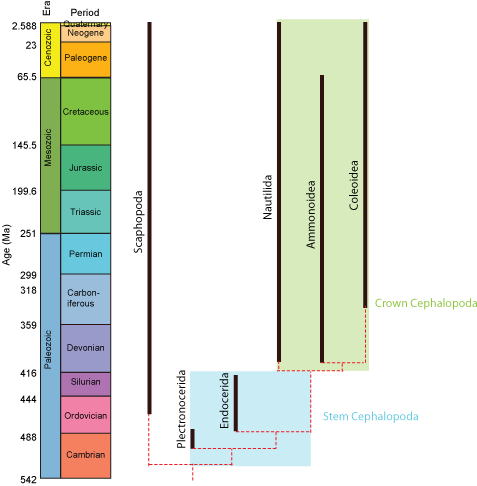
However, we can go beyond simply talking about crown- and stem-groups. For example, Amniota is
defined as "the concestor of Mammalia and Reptilia, and all of its descendants." And Dinosauria is "the concestor
of Iguanodon and Megalosaurus, and all of its descendants.". So any taxon that comes from the node
(joining point) that links Megalosaurus and Iguanodon is a member of Dinosauria (i.e., is a dinosaur); and
anything that lies outside that node is NOT a dinosaur. So in the cladogram below:
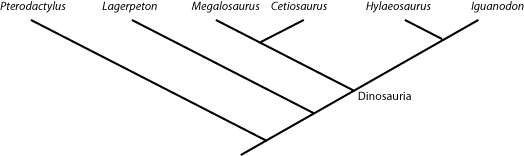
Cetiosaurus, Hylaeosaurus (and, by definition, Megalosaurus and Iguanodon) are all dinosaurs,
but Lagerpeton and Pterodactylus are not.
Names coined in this fashion are said to have node-based taxonomic definitions. These have the form "the concestor of A and B and all of its descendants" or more generally "the least inclusive clade containing A and B". (A and B are said to be the specifiers or anchor taxa.)
Names can also be be defined another way: by sister group relationship rather than by concestry. For example, the two major branches of Dinosauria are Saurischia and Ornithischia. The former is defined as "Megalosaurus and all taxa sharing a more recent common ancestor with Megalosaurus than with Iguanodon", and the latter is defined as "Iguanodon and all taxa sharing a more recent common ancestor with Iguanodon than with Megalosaurus". So Cetiosaurus and Megalosaurus are both saurischians, and Hylaeosaurus and Iguanodon are both ornithischians. These names have branch-based taxonomic definitions. These have the form "A and all taxa sharing a more recent common ancestry with A than with B" or more generally "the most inclusive clade containing A but not B".
Similarly, the clade Dinosauromorpha is defined as Dinosauria and all taxa sharing a more recent common ancestor with
Dinosauria than with Pterodactylus. Showing those relationships (by labeling the name, and by color blobs) onto the
cladogram, we see:
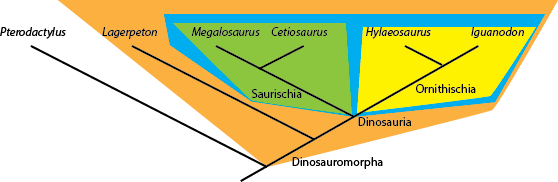
In principle, one can also coin apomorphy-based definitions. These have the form "The clade stemming from the first organism or species to possess apomorphy M as inherited by A" or "the most inclusive clade exhibiting character state M that is synapomorphic with that in A". For example, in the diagram above, Hylaeosaurus is unique in possessing osteoderms (armored bony plates in its skin). One could define a clade "Hylaeosauroidea" with the form "The clade stemming from the first species possessing osteoderms as inherited by Hylaeosaurus". However, most practioners of phylogenetic systems of nomenclature avoid this, as apomorphies almost always exhibit transitional stages, which makes the dividing point operationally difficult to define.
On a cladogram we don't normally see numerical time. Because the nodes on a cladogram represent the presence
of shared common ancestors, they are guide to the relative sequence of divergences. So in the cladogram of
birds and bird-like dinosaurs:
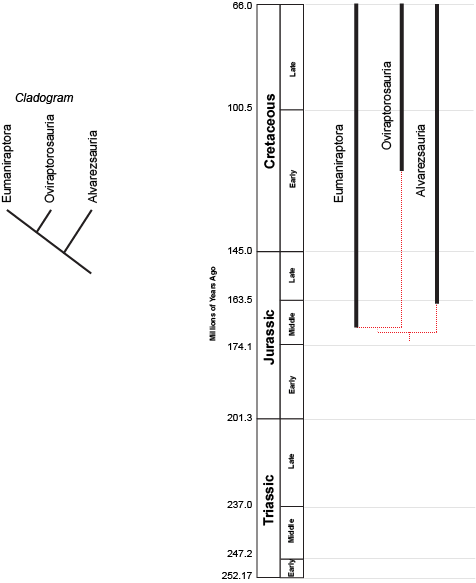
the concestor of Eumaniraptora + Oviraptorosauria must have lived more recently (closer to us in time) than the concestor of (Eumaniraptora + Oviraptorosauria) + Alvarezsauria. (After all, the latter concestor is the ANCESTOR of the (Eumaniraptora + Oviraptorosauria) concestor!) But we can't tell numerical time from a cladogram.
However, we can plot the known stratigraphic ranges (that is, the range between the oldest and youngest specimen) of each taxon. Those are the black lines on the phylogeny to the right above. The dashed red lines represent the cladistic relationships between the taxa. We see that the divergence between Eumaniraptora and Oviraptorosauria had to have happened sometime before the oldest member of these sister taxa (that is, the mid-Middle Jurassic eumaniraptorans). The divergence between Alvarezsauria and the (Eumaniraptora + Oviraptorosauria) had to happen earlier than that. We don't know the exact divergence date unless we actually find the concestors; but we can find the minimum divergence date (in both these cases, the Middle Jurassic).
The phylogeny also shows us other information. We see that Oviraptorosauria is first known from the middle part of the Early Cretaceous. Assuming our cladogram is correct, there should be Middle Jurassic, Late Jurassic, and early Early Cretaceous representatives of the Oviraptorosauria lineage. Since we infer their presence, but haven't confirmed them with direct observations, we refer to this part of the branch as a ghost lineage. Maybe we just haven't found them; maybe they lived in a region that didn't produce fossils; or maybe we have them in collections but currently haven't properly identified them.
There are several reasons why our minimum divergence dates may underestimate the actual time of origin of a clade:
To Syllabus.