Key Points:
•Cladistics (phylogenetic systematics) is a method for approximating the evolutionary relationships among taxa.
•Cladistics works by trying to reconstruct the pattern of common ancestry rather than finding direct ancestor-descendant relationships.
•Not all traits are equally useful for reconstructing phylogenetic relationships: only shared evolutionary transformations help us determine phylogenetic patterns.
•Phylogenetic information can be used as a basis for taxonomy; as a means of inferring missing and ancestral information; and for determining the time of divergence between lineages.
I. The Tree of Life
The most important pattern: the Tree of Life.
Darwin and Wallace demonstrated the reality of
Divergence through Time and Common Ancestry:
This allowed a framework for a new style of systematics:
Darwin recognized that levels of similarity came about because of recency of common ancestry. Bats were more closely related to each other than to other types of mammals because there was a shared common ancestor for all bats more recent in time than a shared common ancestor of bats and other mammals. We recognize this because bats share the basic mammal traits (inherited from the common ancestor of all mammals) PLUS their own special traits (wings, echolocation, etc.) inherited from the common ancestor of all bats.
Darwin advocated a change in Linnaean classification reflect the pattern of common ancestry (he used the more Victorian phrase "propinquity of descent"). He also warned that we can't just use one trait as the basis for all the classification, because of the possibility that trait evolved convergently (independently) in multiple lines. Instead, we have to use the aggregate of multiple traits.
Ernst Haeckel coined the word phylogeny for a "family tree" of Life (or some subset thereof). This is a representation of an hypothesis of the actual pattern of ancestors and descendants. Since all living things are related (at least at some level), there should be one true historical branching Tree of Life. The task, then, is to reconstruct it.
Ideally, one could combine information from living creatures with their ancestors to complete this tree. But there are difficulties:
During the late 19th and much of the 20th Century, paleontologists and other biologists constructed phylogenies based on observed features in the organisms, put them into proper stratigraphic context, and "connected the dots". What was needed was a more rigorous methodology.
II. Cladistics
That method was developed by East German entomologist Willi Hennig
in the middle of the 20th Century. The technique was called phylogenetic systematics, or more commonly cladistics
(from the Greek "klados" ["branch"] for clade, meaning a branch of the Tree of Life).
Hennig recognized that finding direct ancestors in the fossil record would be hard, and demonstrating that the fossil
was a direct ancestor rather than a close relative of that ancestor would be even harder. So instead, his method focused
on estimating the pattern of shared common ancestry. In order to make that estimation, cladistics looks at the
distribution of derived characters [evolutionary specializations]. Here is how it works. Below is a grouping
of some modern animals:
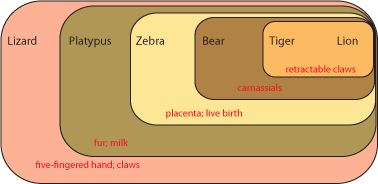
Each set contains multiple taxa. Each set is characterized by derived characters not seen in the taxa outside that
set. For example, the lizard shares with all the others the presence of a five-fingered hand and claws.
The platypus and all the others except the lizard shares fur and milk. Since lizards (and alligators and frogs and
fish and many other animals not shown here) do not have fur and milk, it is most likely explained by having evolved in the
common ancestor of platypus, zebra, bear, tiger, and lion AFTER that ancestor had diverged from the common ancestor
of lizard, platypus, zebra, bear, tiger, and lion. In other words, platypus, zebra, bear, tiger, and lion most likely
had a more recent common ancestor with each other than any did with lizards.
It is generally easier to show this pattern using a simplified stick figure called a cladogram than the set diagram.
Below is the cladogram showing the same information as the sets above:
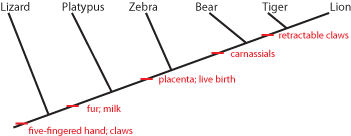
The little red "tick marks" show that the derived character state appears at that point of the cladogram, and is passed
on upwards. To show what that means, see the following:

So the common ancestor of zebra, bear, tiger, and lion evolved a placenta and live birth, and passed it on to its descendants.
And the common ancestor of bear, tiger, and lion (but NOT zebra) had its molars evolve into carnassials (shearing teeth), and
passed that trait on.
Not all character states are useful in figuring out evolutionary relationships. Primitive (or ancestral) states do not help. For example, both lizard and platypus lay shelled eggs rather than have a placenta and live birth. Does that mean that these two animals shared a more recent common ancestor with each other than either did with the other animals here? NO! In fact, having a shelled egg is the ancestral state for a larger (more inclusive) group of animals, including birds, crocodilians, turtles, snakes, echidnas, etc. Having a shelled egg is simply the condition (state) of all advanced terrestrial vertebrates. However, fur and milk are derived states not found in terrestrial vertebrates other than mammals, and indicate a common ancestor to these animals not found in lizards, birds, crocodilians, turtles, etc.
Similarly, sharing a five-fingered hand doesn't unite the lizard, platypus, and bear (for example) exclusive of the zebra. A five-fingered hand is the primitive condition, and will be passed on that way unless it evolves into something else.
(NOTE: a character can be "primitive" at one level, but "derived" in a different context. In this cladogram, retractable claws is a derived feature that shows us that lions and tigers share a more recent common ancestor with each other than with anything else on this cladogram. However, if we were trying to figure out the relationship between lions, tigers, jaguars, and leopards, than retractable claws would be a primitive character shared by all of them, and so useless.)
Zebras have only a single finger on each hand. This is a unique derived character. Although unique derived characters might be useful in understanding how that animal lived, they don't help us figure out where they fit in the cladogram, since those traits evolved after the ancestor of (in this case) zebras diverged from any shared ancestor of any of the other animals on the list. (However, if we added horses or donkeys to this cladogram, a single finger on each hand would be a good shared DERIVED character, and help us figure out that horses and donkeys share a common ancestor not shared with bears, lions, tigers, etc.).
Convergent characters could actually mislead us. For example, tigers and zebras both have stripes, whereas the other animals on this list don't. Does that mean that tigers and zebras share a common ancestor not shared by the other animals? We might think so at first, but the shared presence of carnassials in bears, lions, and tigers (but not zebras) and of retractable claws in lions and tigers (but not zebras) indicate a different pattern of ancestry. Convergent characters can be very annoying, because they might cause us to make the wrong estimation of the cladogram.
An additional type of character (not seen here) are reversals: when an ancestor had a derived condition, but the trait evolved "back" to the primitive condition. For example, snakes are descendants of typical legged lizards. But snakes have evolved the loss of limbs, so that they resemble the limbless ancestor of vertebrates. For another, while the common ancestor of living birds could fly, but ostriches, rheas, cassowaries, etc. have reverted to the primitive flightless state. Reversals that are unique characters do not help in estimating evolutionary relationships, but ones which are shared might. In general, though, reversals (like convergent characters) are annoying and are a major source of error for our analyses.
So, in review:
How do we tell which states (conditions) of a character are ancestral, and which are derived? One common method is to look at outgroups: more distant relatives of the taxa in question. Looking at fish, amphibians, turtles, lizards and snakes, crocodilians, and birds show that no placenta is the primitive condition among vertebrates, and that the placenta of some mammals is a derived condition. (It is best to look at multiple outgroups.)
How do we actually build the tree? In order to start a phylogenetic analysis, we construct a dataset of the characters and taxa we want to study, and code each taxon as to what character state it has. (These characters (features) could be DNA sequences, behaviors, and so on: for fossils, we are stuck with physical features for the most part!) We then reconstruct the different possible branching sequences, and "map" the distribution of the derived character states onto the cladogram. We count each time a character state changes, and sum up all the changes. The cladogram(s) with the smallest number of changes is/are the one(s) we prefer: it/they are the simplest possible explanation for the observations (this is known in Science as the principle of parsimony). Future studies (with new characters, new taxa, or both) might support our first estimations, or they might give us new results.
Note that the number of possible cladograms increases with the number of taxa studied. Because of this, we use computer software to estimate our cladograms: it would take more time than is humanly possible otherwise!!
III. Tree-Based Thinking
You won't be creating cladograms in this course, but you WILL be reading them. One of the most important parts of an
evolutionary perspective on the history of life is to stop thinking about "kinds" or "types" of organisms as set distinct
things. Instead, use tree-based thinking: remember that all organisms past and present are part of the Tree of Life.
So be able to read a cladogram to understand the shared pattern of ancestry and the passing on of evolutionary specializations.
Here are some useful guides:
A. A Cladogram is a Stick-Figure Version of an Hypothesis of the Pattern of Shared Common Ancestry
Remember we could convey the same information with set diagrams. It is NOT a "family tree" in the traditional
context, in that it doesn't show ancestors and descendants. For example, in the cladograms shown above, zebras are
NOT the ancestor of bears, tigers, and lions; bears are not the ancestors of tigers and lions, and tigers are not
the ancestors of lions. Instead, it DOES show that lions and tigers share a common ancestor not shared with bears; that
lions, tigers, and bears share a common ancestor not shared with zebras; and so on.
Also, any cladogram is just a small subsample of the total diversity of life. There are many animals not shown on the cladogram above that descend from (for example) the common ancestor of
B. The Important Information of a Cladogram is the Branching Pattern
In a cladogram, it is the branching relationships which are important, not the "right
to left"/"top to bottom" order. As long as two cladograms contain the same branching relationships,
and do not have any contradictory branching relationships, they are equivalent:
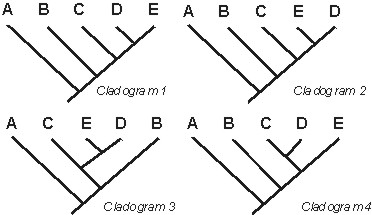
In the above cases,cladograms 1-3 are all equivalent: they represent the same information. However, cladogram 4 is not equivalent to the other three:
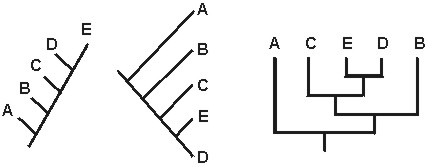
C. Relationships (and Groupings) are based on Recency of Common Ancestry, Not Overall Similarity
Darwin, Hennig, and their followers advocate the use of common ancestry, rather than overall similarity, as the basis for classification. The reason?
Overall similarity might represent common ancestry if the organisms are similar because of shared derived characters.
But overall similarity might also simply represent the shared inheritance of primitive traits.
For example, below is the cladogram for the living members of Tetrapoda, the clade (group) of terrestrial vertebrates:
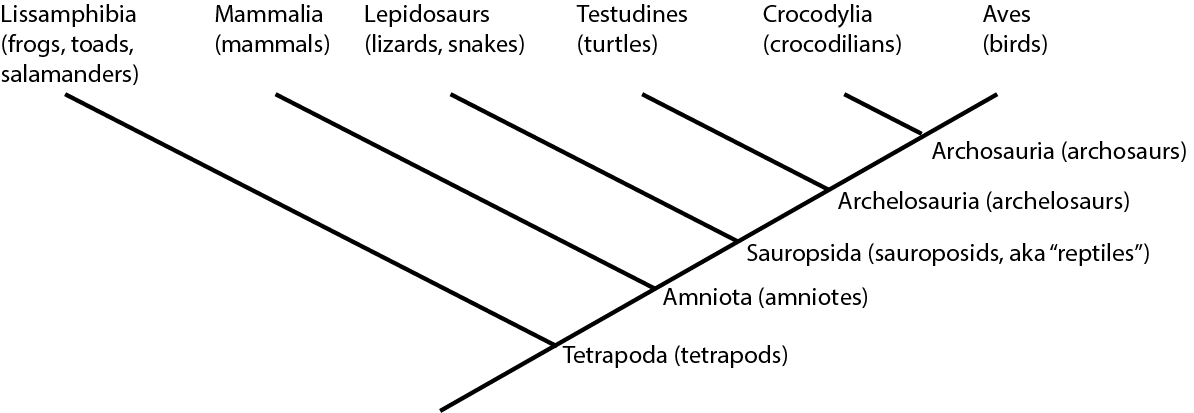
On a cladogram, a name refers to the entire clade: an ancestor plus ALL of its descendants. (Another name for
"clade" is "monophyletic group" [monophyly = one branch].)
We could also write out this information like an outline: Tetrapoda
So:
Traditionally, crocodilians would be thought to be "more closely related" to lepidosaurs and turtles than to birds, because of the
overall similarity. But this overall similarity was simply shared primitive features. Instead, as we will see, crocodilians
and birds actually share far more derived features with each other than either does with lepidosaurs or turtles. Thus,
birds and crocodilians are more closely related to each other than either are to lepidosaurs or turtles. (To
use the technical term, Aves and Crocodylia are each other's sister group.):

In fact, lepidosaurs are no more closely related to crocodylians than they are to birds! They simply resemble crocodilians more because the bird lineage has gone through many more spectacular evolutionary transformations!
Note that the traditional version of "Reptilia" (= Testudines, Lepidosauria, Crocodylia, but not Aves) is NOT a monophyletic group. Instead, it is what is called a paraphyletic group: an ancestor, but NOT all of its descendants.
You can see the distinction here:
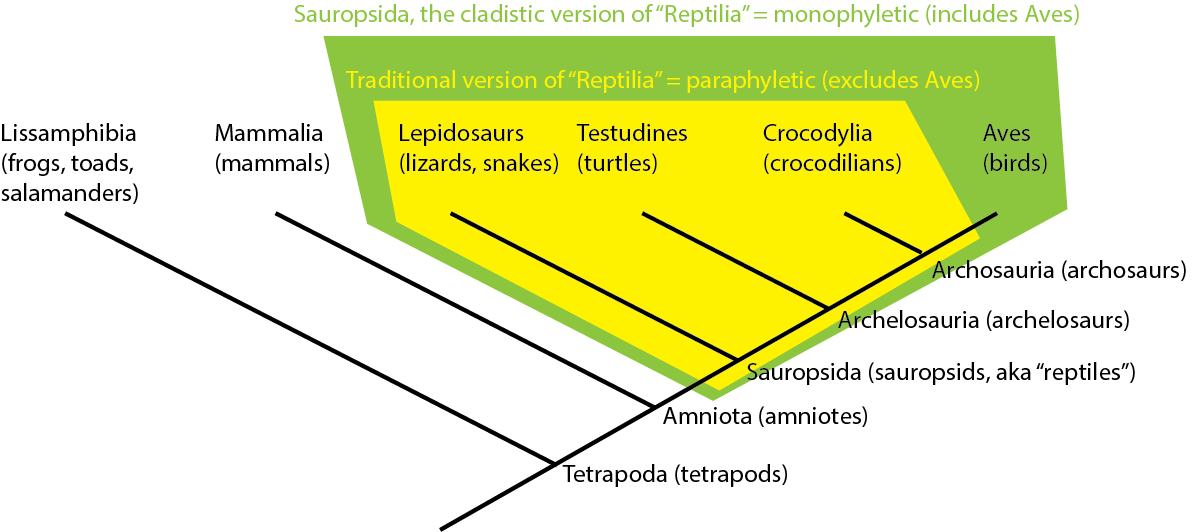
Many traditional Linnaean groups have turned out to be paraphyletic groupings (Pisces; Invertebrata; Fissipedia; etc.).
However, because cladistic taxonomy uses only clades (monophyletic groups), we no longer use these paraphyletic taxa.
NOTE: In this course we will use the name "Sauropsida" for the monophyletic group (clade) containing Testudines, Lepidosauria, Crocodylia, Aves, and all taxa closer to these than to Mammalia; however, some still use "Reptilia" for this same clade.
D. Phylogenetic Taxonomic Names are Defined by Patterns of Relationships
Paleontologists and other biologists have started to use phylogenetic definitions for taxa: that is, they specify a
particular relationship and any taxon that qualifies under that relationship is member of that clade.
Biologists have coined the word concestor for most recent common ancestor.
While the chances of finding the concestor of any two (or more) clades is slim, we can recognize that there did exist
such a species somewhere in time. This recognition gives us a way of defining taxa. For example, Amniota is
defined as "the concestor of Mammalia and Sauropsida, and all of its descendants." And Dinosauria is "the concestor
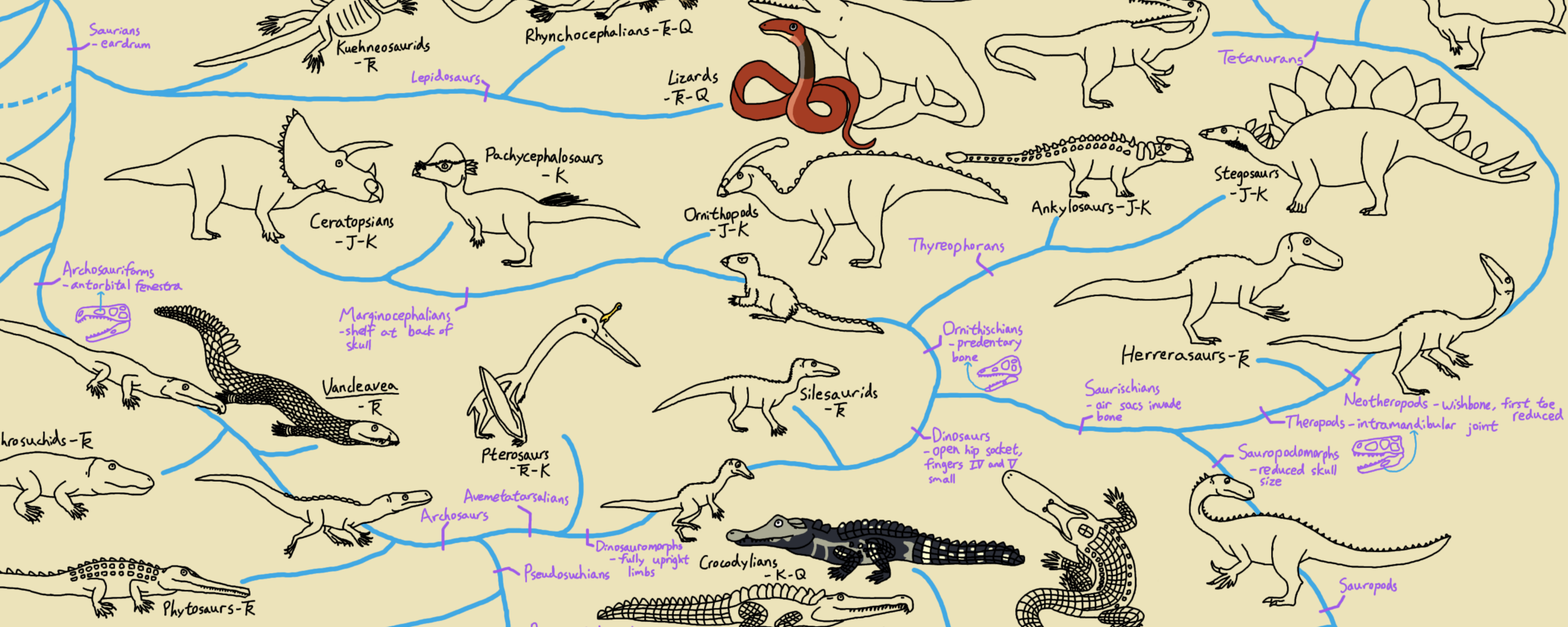
of Iguanodon, Diplodocus, and Megalosaurus, and all of its descendants.". So any taxon that comes from the node (joining point) that links Megalosaurus, Diplodocus, and Iguanodon is a member of Dinosauria (i.e., is a dinosaur); and anything that lies outside that node is NOT a dinosaur. So in the cladogram below:
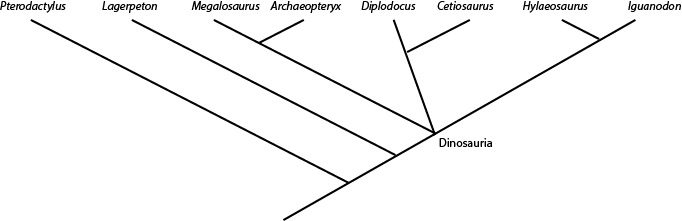
Archaeopteryx, Cetiosaurus, Hylaeosaurus (and, by definition, Megalosaurus, Diplodocus, and Iguanodon) are all dinosaurs, but Lagerpeton and Pterodactylus are not.
Names can also be be defined another way: by sister group relationship rather than by concestry. For example, the three major branches of Dinosauria are Theropoda, Sauropodomorpha, and Ornithischia. The first is defined as "Megalosaurus and all taxa sharing a more recent common ancestor with Megalosaurus than with Diplodocus or Iguanodon"; the middle one is "Diplodocus and all taxa sharing a more recent common ancestor with Diplodocus than with Megalosaurus or Iguanodon"; and the last (not surprisingly) is defined as "Iguanodon and all taxa sharing a more recent common ancestor with Iguanodon than with Megalosaurus or Diplodocus". So Cetiosaurus and Diplodocus are both sauropodomorphs, Megalosaurus and Archaeopteryx are both theropods, and Hylaeosaurus and Iguanodon are both ornithischians.
Similarly, the clade Dinosauromorpha is defined as Dinosauria and all taxa sharing a more recent common ancestor with
Dinosauria than with Pterodactylus. Showing those relationships (by labeling the name, and by color blobs) onto the
cladogram, we see:
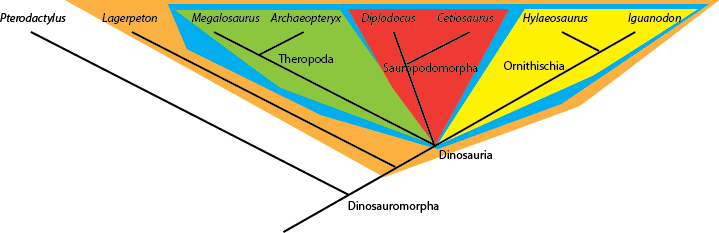
E. We Can (Sometimes) Use Cladograms To Predict Missing Information
No fossil is complete, so we can't always directly tell what character state was present in a given species. But we
can sometimes us the phylogenetic position of a fossil to help us estimate what that state was. Below is a cladogram
of tyrannosauroids (tyrant dinosaurs), with the number of fingers on the hand listed:
:
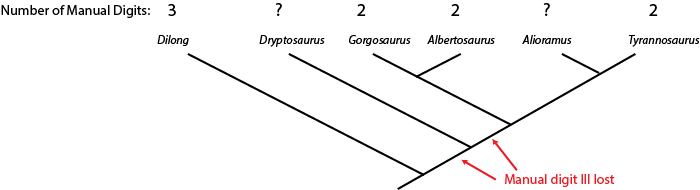
The hand is not yet known for Alioramus or Dryptosaurus. However, we know that Dilong has the
ancestral condition of a three fingered manus (shared with many other carnivorous dinosaur groups), and that Gorgosaurus,
Albertosaurus, and Tyrannosaurus all share the evolutionary specialization "loss of manual digit III" (meaning
that they had a two-fingered hand). The simplest explanation is that the concestor of these three dinosaurs had already lost
manual digit III, and passed on this derived trait to the different descendant lineages.
Because Alioramus (based on other evidence) is ALSO a descendant of that concestor, we predict that it too had a two fingered hand. To assume it had a three fingered hand would mean assuming an evolutionary change (in this case, a reversal) for which we had no direct evidence. The simplest explanation for our current observations is that Alioramus was a two-fingered dinosaur.
But things are unclear for Dryptosaurus. It might be that the loss of manual digit III occurred after the ancestor of Dryptosaurus diverged from the concestor of Alberosaurus through Tyrannosaurus: if that were the case, Alectrosaurus would have a three fingered hand. OR it might be that the loss of manual digit III happened before the Dryptosaurus lineage and the Albertosaurus + Tyrannosaurus lineage had diverged: in that case, Dryptosaurus was a two fingered dinosaur. So at present, things remain ambiguous for this taxon.
F. We Sometimes Get Ambiguous Results
Because of missing information, convergence, and reversals, there can be confusion as to the actual pattern of the Tree of
Life. It is often the case that multiple different cladograms are equally well supported ("equally parsimonious"). In
those cases, we can list all of them separately, or we can show a "consensus cladogram" that shows the ambiguity. This is
seen by the presence of a polytomy: a node with more than two branches coming out of it. Below are three different
equally simple cladograms for the sauropsids, and the consensus cladogram below:
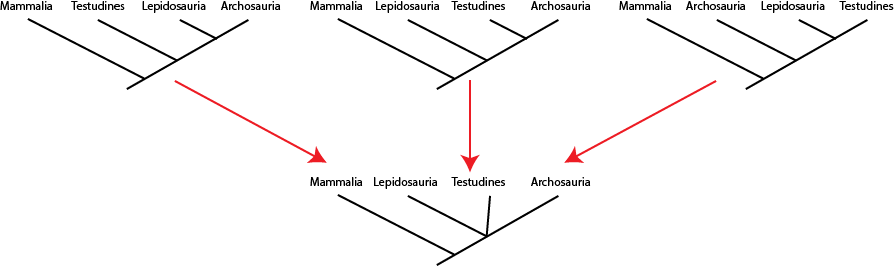
Note the polytomy in the consensus cladogram. (The origin of turtles is outside the main scope of this course, but remains one of the least-resolved part of the vertebrate family tree.)
G. We Can Combine Cladograms and Stratigraphy
On a cladogram we don't normally see numerical time. Because the nodes on a cladogram represent the presence
of shared common ancestors, they are guide to the relative sequence of divergences. So in the cladogram of
bird-like dinosaurs to the left below:
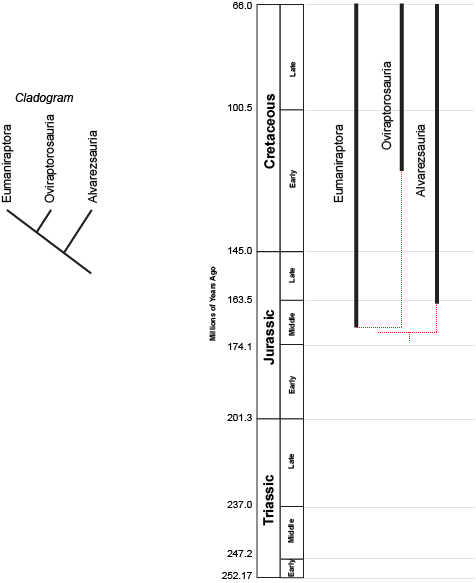
the concestor of Eumaniraptora + Oviraptorosauria must have lived more recently (closer to us in time) than the concestor of
(Eumaniraptora + Oviraptorosauria) + Alvarezsauria. (After all, the latter concestor is the ANCESTOR of the (Eumaniraptora + Oviraptorosauria) concestor!) But we can't tell numerical time from a cladogram.
However, we can plot the known stratigraphic ranges (that is, the range between the oldest and youngest specimen) of each taxon. Those are the black lines on the phylogeny to the right above. The dashed red lines represent the cladistic relationships between the taxa. We see that the divergence between Eumaniraptora and Oviraptorosauria had to have happened sometime before the oldest member of these sister taxa. The divergence between Alvarezsauria and the (Eumaniraptora + Oviraptorosauria) clade had to happen even earlier than that. We don't know the exact divergence date unless we actually find the concestors; but we can find the minimum divergence date (in both these cases, the Middle Jurassic).
The phylogeny also shows us other information. We see that the oldest known members of Eumaniraptora are from the Middle Jurassic, that the oldest known members of Oviraptorosauria date from the mid-Early Cretaceous; that the oldest known members of Alvarezsauria date from near the Middle-Late Jurassic boundary; and that all these groups die out at the end of the Cretaceous.
We can also see that there are still plenty of dinosaurs to find! Assuming our cladogram is correct, there should be Middle Jurassic, Late Jurassic, and early Early Cretaceous representatives of the Oviraptorosauria-lineage (these wouldn't be any of the known oviraptorosaurs themselves, but dinosaurs sharing a more recent common ancestor with them than with eumaniraptorans). It might be that we already have these fossils but don't recognize their phylogenetic position (indeed, some think that the Jurassic Scansoriopterygidae are early oviraptorosaurs), or we simply may not have found them yet.
(Just so you know, I went out of my way to choose a case where the branching events on the cladogram do not match up well with the stratigraphic order of the appearance of the clades. As you will see in later lectures, these patterns generally match up pretty well!)
In the rest of the course, you will find both simplified cladograms (showing major taxa and some major shared derived characters) and more detailed phylogenies (showing additional taxa, as well as their stratigraphic ranges). Please make certain that you are able to read and understand these.
To Next Lecture.
To Previous Lecture.
To Lecture Notes.